Normalization workflow with script
In this example we transform raw time-resolved plant phenotypes into normalized and averaged results, based on the metadata labels included in the dataset.
click here to download the full script (shown at the bottom of this page).
click here to download the sample dataset. This file can be dragged directly into the OLIVER window.
1. Normalize experiment samples against control samples
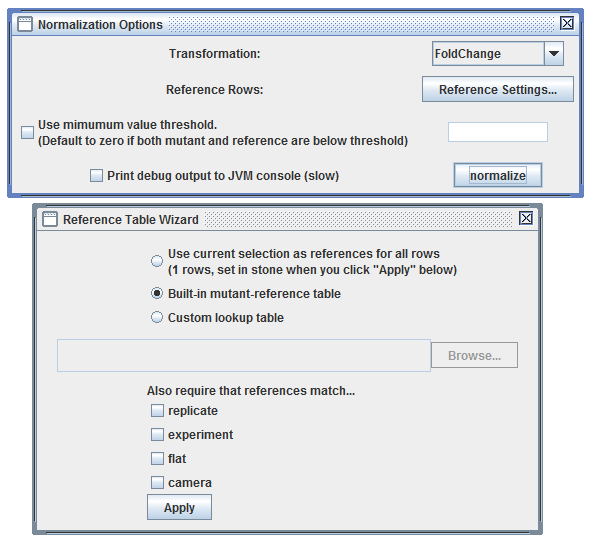
User Interface: load a heatmap and click "Tools", "Create Normalized Heatmap", "By Reference Rows..."

|
|
|
Launching normalization dialog using the heatmap "Tools" menu |
The "Reference Table Wizard" is launched by clicking "Reference Settings..." |
Scripting: create a ReferenceTable object which relates each experiment group in our dataset. We use that to create a HeatmapNormalizer object which will generate a new heatmap when we call it's buildNormalizedMap() function.
refTable = new ReferenceTable( phi2_raw, "Flat" )
phi2_lfc = new HeatmapNormalizer( phi2_raw ).buildNormalizedMap( LogFoldChange, refTable )
- Since this sample dataset uses standard experiment/control row labels, OLIVER will automatically recognize them.
"Flat"is a metadata field included in the row labels for our data. Adding it here indicates that we only want to compare experiment samples with controls with the same "Flat".LogFoldChangeis a predefined constant, representing one type of normalization. Other acceptable types includePercentDifferenceandStandardScore.
2. Replace infinite values with blanks
This is a special step that may be useful for making sense of certain normalization results (e.g. if the control group has a value of zero which is used as a denominator)
User Interface: Not supported.
Scripting: Use a heatmap's .replaceInfiniteValues() function, and specify the new value to replace infinities. We represent an empty cell using NaN ("Not a Number")
phi2_lfc.replaceInfiniteValues( NaN )
3. Separate experiment results from controls
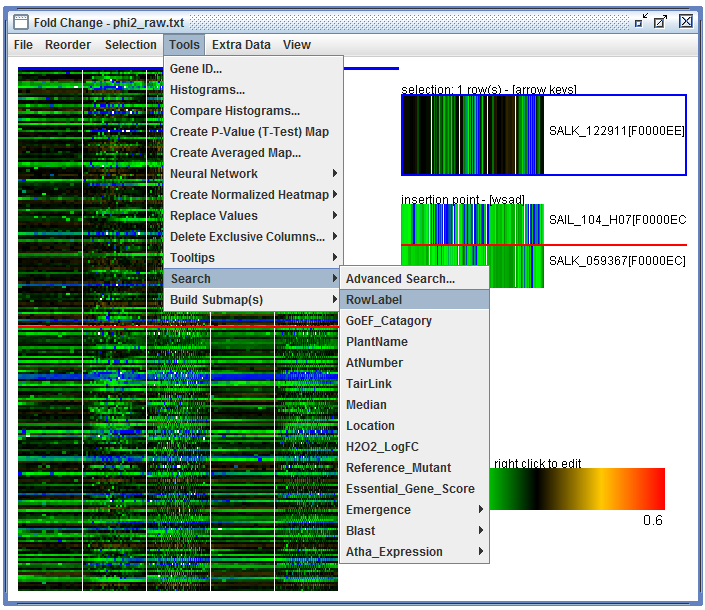
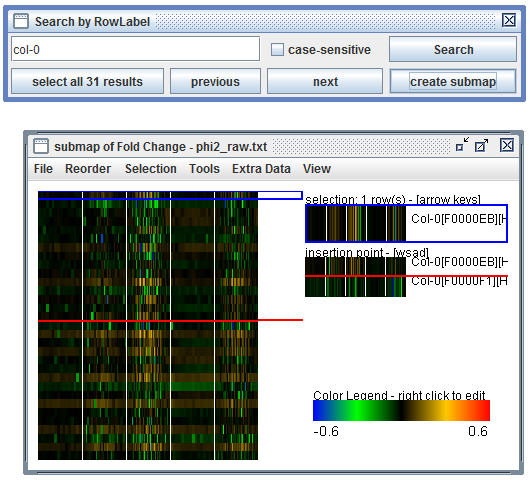
User Interface: In a heatmap click "Tools", "Search", "Row Label", enter a search term, "Search", "select all results", "create submap". Then "Selection", "Invert selection" and then "Selection", "Submap from Selection"
|
|
|
Launching search dialog using the heatmap "Tools" menu |
After entering a search term, click "Search", "select all results", and "create submap". You can invert the selection before making the submap using "Selection", "Invert Selection". |
Scripting: Use the phi2_lfc heatmap's searchRowLabels() function to create a new variable sel_wt, containing the set of searched rows. Then use that set of rows in conjuction with the getSubmap() and invert() functions of phi2_lfc to create two submaps.
sel_wt = phi2_lfc.searchRowLabels( "col-0" )
phi2_lfc_wt = phi2_lfc.getSubMap( sel_wt )
phi2_lfc_mt = phi2_lfc.getSubMap( phi2_lfc.invert( sel_wt ) )
4. Average replicates into single rows
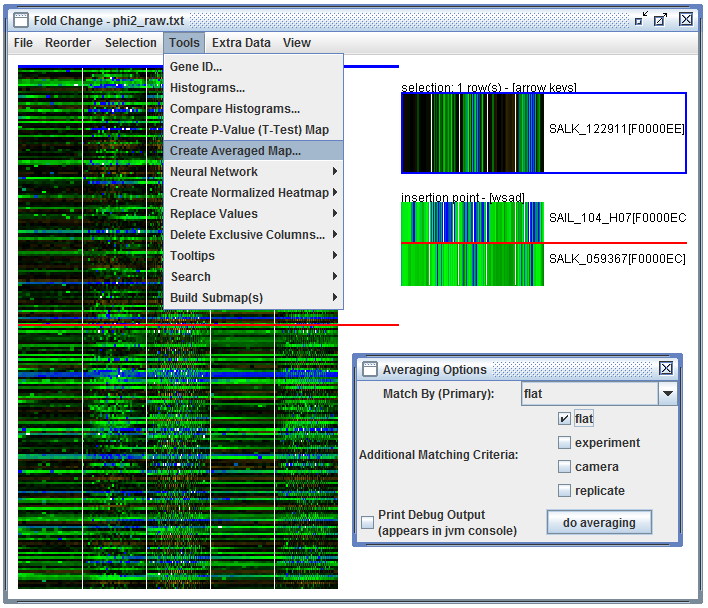
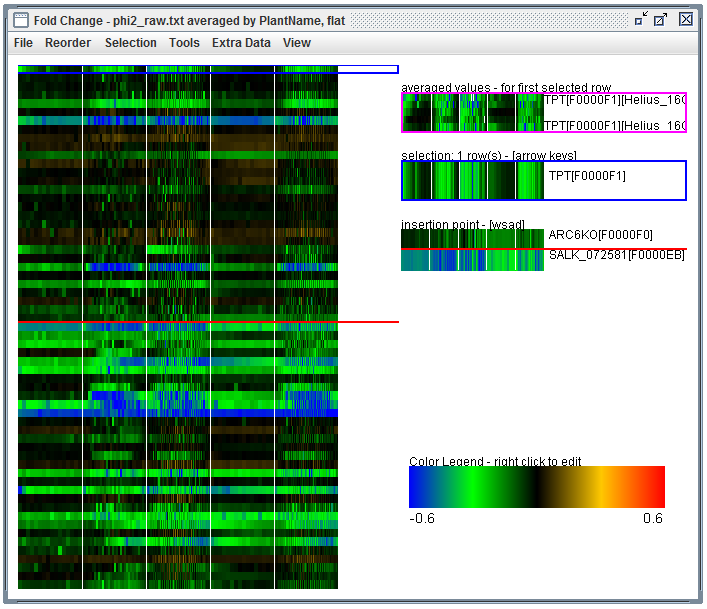
User Interface: In a heatmap and click "Tools", "Create Averaged Map...". Then match by "PlantName", check "flat", and click "do averaging".
|
|
|
averaging dialog. Launched using the heatmap "Tools" menu |
Averaged results. |
Scripting: create a HeatmapAverager object and use it's getAveragedMap() function, providing the fields that should be matching for each set of rows that should be averaged.
phi2_lfc_mt_avg = new HeatmapAverager( phi2_lfc_mt ).getAveragedMap( "PlantName", "flat" )
phi2_lfc_wt_avg = new HeatmapAverager( phi2_lfc_wt ).getAveragedMap( "PlantName", "flat" )
5. Average results with matching meta-data
User Interface: In a heatmap and click "Tools", "Create Averaged Map...". Then match by "AtNumber" and click "do averaging".
Scripting: as in the previous section, use HeatmapAverager and getAveragedMap()
phi2_lfc_mt_avg_atg = new HeatmapAverager( phi2_lfc_mt_avg ).getAveragedMap( "AtNumber" )
6. Replace missing data with zeroes
User Interface: Not supported.
Scripting: Use the heatmap function replaceValues() to select values that should be replaced (in this example we do that be defining a new function that returns "True" for NaNs). The second parameter passed to the replaceValues() function will be used as the replacement.
phi2_lfc_mt_avg.replaceValues( function(x){ return x=NaN; }, 0 )
phi2_lfc_wt_avg.replaceValues( function(x){ return x=NaN; }, 0 )
phi2_lfc_mt_avg_atg.replaceValues( function(x){ return x=NaN; }, 0 )
7. Delete some non-relevant rows
User Interface:
- click "Tools", "Search", "Row Label"
- Type "null" and click "Search", "Select all results"
- click "Selection", "Delete Selected Rows"
Scripting: use a heatmap's .deleteRows() function in combination with .searchRowLabels()
phi2_lfc_mt_avg.deleteRows( phi2_lfc_mt_avg.searchRowLabels( "null" ) )
phi2_lfc_wt_avg.deleteRows( phi2_lfc_wt_avg.searchRowLabels( "null" ) )
phi2_lfc_mt_avg_atg.deleteRows( phi2_lfc_mt_avg_atg.searchRowLabels( "null" ) )
8. Display results
User Interface: not appicable - results are automatically displayed
Scripting: use .setTitle() on a heatmap to specify the title to use for displaying it. Then use GLOBAL.display() to show any number of heatmaps. This way you can choose not to clutter the screen with intermediate results
phi2_lfc_mt_avg.setTitle( "LogFoldChanged Phi2 - mutants averaged by PlantName/Flat" )
phi2_lfc_wt_avg.setTitle( "LogFoldChanged Phi2 - wildtypes averaged by PlantName/Flat" )
phi2_lfc_mt_avg_atg.setTitle( "LogFoldChanged Phi2 - mutants averaged by AtNumber" )
GLOBAL.display( phi2_lfc_mt_avg, phi2_lfc_wt_avg, phi2_lfc_mt_avg_atg )
Complete script and sample data
click here to download the full script (shown below).
click here to download the sample dataset. This file can be dragged directly into the OLIVER window.
//phi2
//1) compute LFC by Tools --> Create normalized map --> Transformation = LogFolfChange, ReferenceRows = lookup_table, FilterReference = flat
//2) replace -Infinity and Infinity by NaN if any (there are many NaNs, but no worry)
//3) split result to two files: mutant (mt) and col-0 (wt)
//4) compute the avg LFC by Tools --> Create averaged map -->MatchBy = SalkID, AdditionalMatchingCriteria = flat
//5) compute atg averaged LFC by Tools --> Create averaged map -->MatchBy = AtNumber
//6) replace all NaN by 0 because logged fold change=NaN is only caused by the divided by 0 error (caused by replacing negative or small phenotype with 0)
//7) delete the row with its name "null"
//8) display results:
// multi-allPhi2-3-day-lfc2-mt-avg-atg-abn.txt
// multi-allPhi2-3-day-lfc2-wt-avg.txt
//the comment below shows how you could load data from your hard drive
//phi2_raw = GLOBAL.loadHeatmapFromPath("C:\\Users\\Guest\\Documents\\Linda-final-expc\\phi2\\allPhi2.txt")
//1
refTable = new ReferenceTable( phi2_raw, "Flat" )
phi2_lfc = new HeatmapNormalizer( phi2_raw ).buildNormalizedMap( LogFoldChange, refTable )
//2
phi2_lfc.replaceInfiniteValues( NaN )
//3
sel_wt = phi2_lfc.searchRowLabels( "col-0" )
phi2_lfc_wt = phi2_lfc.getSubMap( sel_wt )
phi2_lfc_mt = phi2_lfc.getSubMap( phi2_lfc.invert( sel_wt ) )
//4
phi2_lfc_mt_avg = new HeatmapAverager( phi2_lfc_mt ).getAveragedMap( "PlantName", "flat" )
phi2_lfc_wt_avg = new HeatmapAverager( phi2_lfc_wt ).getAveragedMap( "PlantName", "flat" )
//5
phi2_lfc_mt_avg_atg = new HeatmapAverager( phi2_lfc_mt_avg ).getAveragedMap( "AtNumber" )
//6
phi2_lfc_mt_avg.replaceValues( function(x){ return x=NaN; }, 0 )
phi2_lfc_wt_avg.replaceValues( function(x){ return x=NaN; }, 0 )
phi2_lfc_mt_avg_atg.replaceValues( function(x){ return x=NaN; }, 0 )
//7
phi2_lfc_mt_avg.deleteRows( phi2_lfc_mt_avg.searchRowLabels( "null" ) )
phi2_lfc_wt_avg.deleteRows( phi2_lfc_wt_avg.searchRowLabels( "null" ) )
phi2_lfc_mt_avg_atg.deleteRows( phi2_lfc_mt_avg_atg.searchRowLabels( "null" ) )
//8
phi2_lfc_mt_avg.setTitle( "LogFoldChanged Phi2 - mutants averaged by PlantName/Flat" )
phi2_lfc_wt_avg.setTitle( "LogFoldChanged Phi2 - wildtypes averaged by PlantName/Flat" )
phi2_lfc_mt_avg_atg.setTitle( "LogFoldChanged Phi2 - mutants averaged by AtNumber" )
GLOBAL.display( phi2_lfc_mt_avg, phi2_lfc_wt_avg, phi2_lfc_mt_avg_atg )